- Research article
- Open access
- Published:
QligFEP: an automated workflow for small molecule free energy calculations in Q
Journal of Cheminformatics volume 11, Article number: 26 (2019)
Abstract
The process of ligand binding to a biological target can be represented as the equilibrium between the relevant solvated and bound states of the ligand. This which is the basis of structure-based, rigorous methods such as the estimation of relative binding affinities by free energy perturbation (FEP). Despite the growing capacity of computing power and the development of more accurate force fields, a high throughput application of FEP is currently hampered due to the need, in the current schemes, of an expert user definition of the “alchemical” transformations between molecules in the series explored. Here, we present QligFEP, a solution to this problem using an automated workflow for FEP calculations based on a dual topology approach. In this scheme, the starting poses of each of the two ligands, for which the relative affinity is to be calculated, are explicitly present in the MD simulations associated with the (dual topology) FEP transformation, making the perturbation pathway between the two ligands univocal. We show that this generalized method can be applied to accurately estimate solvation free energies for amino acid sidechain mimics, as well as the binding affinity shifts due to the chemical changes typical of lead optimization processes. This is illustrated in a number of protein systems extracted from other FEP studies in the literature: inhibitors of CDK2 kinase and a series of A2A adenosine G protein-coupled receptor antagonists, where the results obtained with QligFEP are in excellent agreement with experimental data. In addition, our protocol allows for scaffold hopping perturbations to identify the binding affinities between different core scaffolds, which we illustrate with a series of Chk1 kinase inhibitors. QligFEP is implemented in the open-source MD package Q, and works with the most common family of force fields: OPLS, CHARMM and AMBER.
Introduction
Calculating physicochemical properties of drug like molecules, such as the solvation free energies or the binding affinities for biological targets, has been a longstanding challenge to computational chemists. The recent increase in computational power and the improvement of force fields to accurately represent the physical properties of drug-like molecules [1,2,3,4,5,6] have set the grounds for molecular dynamics (MD) based methods to be routinely used to address these issues. Particularly, the rigorous free energy perturbation (FEP) methodology, despite being implemented in the framework of biochemical simulations decades ago [7], has recently gained recognition in the optimization of chemical modulators of biological targets, both in academia and pharmaceutical industry pipelines [8]. Besides the technical advances underlying the success of this approach, automatization tools are key to turn an otherwise time-consuming and error prone process into a systematically applicable tool [9, 10].
In contrast to plain, unbiased MD simulations, FEP simulations drive the conversion (or perturbation) of one molecule into another. If we think of two drug-like molecules for which the binding affinity for a given protein is compared, the FEP simulation will connect them through a series of unphysical intermediates [11]. If we translate this to the more typical exploration of a congeneric series of ligands around a given lead molecule, this can be seen as a system of nodes interconnected by edges, each of them representing a perturbation between the pair of molecules involved. If the collection of nodes is finite, e.g. in the case of amino acid mutations, one can define perturbation libraries for all possible permutations in the 20 × 20 matrix, and systematically apply this to evaluate ligand binding affinity for single-point protein mutants [12,13,14]. However, for drug like molecules the number of nodes considerable increases and the chemical space covered becomes several orders of magnitude larger (in the order of 1033 for all molecules adhering to Lipinski’s rule of five [15, 16]), making it impossible to predesign perturbation libraries for ligands. This introduces a bottleneck in the application of FEP simulations in real drug design projects, as the manual setup required is tedious, time consuming and prone to errors. This problem has been recognized by both the academia and industry and recently some protocols were proposed for a more efficient, automated setup of FEP simulations [9, 10, 17,18,19]. The underlying algorithms in those implementations are based on the calculation of maximum common substructure (MCS) between molecule pairs, and perturbations are performed along the edges of those connected nodes that preserve maximum similarity. Additionally, the selection of nodes can a posteriori be refined by a cycle closure analysis, which allows assessing the statistical errors by considering a given node more than once [17].
Next, a decision needs to be made on how to represent the edges, i.e. the perturbation pathway connecting the two molecules. Two major approaches have been proposed, single or dual topology [20]. A single topology consists of a one-to-one atom mapping between the two end point molecular systems. Here non-equivalent atoms in either of the end-states are represented by dummy particles (without non-bonded interactions) and only one set of coordinates need to be dealt with. This approach is particularly useful when changes between the molecules are small. However, breaking (or making) bonds becomes necessary when the bond topology changes more drastically (e.g. the opening of a ring structure in a molecule), which has been shown to significantly hamper convergence [21]. In such cases, a dual topology representation can be adopted. Here, the molecular entities from both nodes are simultaneously represented by their own set of Cartesian coordinates. In each end state of the simulation one of the molecules has its full interactions turned on, whereas the other is turned off (all dummy atoms). Since the total potential energy is a linear combination of the potential energy of each of the two states over the whole perturbation, the two molecules do not interact and the surrounding system sees a mixture of these two states [9, 22].
In this work, we describe an automated protocol to setup ligand perturbations utilizing a dual topology approach called QligFEP, which is implemented to interact with the open-source MD package Q. This software is specifically tailored to perform different types of free energy simulations, such as linear interaction energy (LIE), empirical valence bond (EVB) and FEP [23, 24]. Simulations are run in an explicit spherical droplet of water around the area of interest, i.e. the binding site. For free energy calculations, this has various advantages as compared to the more commonly used periodic boundary conditions (which are also implemented in Q), such as: (1) the absence of artificial periodicity in the finite system; (2) the possibility to calculate all interactions within the droplet accurately either directly or by utilizing multipole expansions for long-range interactions [23, 25]; (3) Focusing on the binding site whilst ‘cutting out’ the envelope allows to perform many multiple independent simulations, thus increasing the precision of the calculated free energies since the motions distal from the region of interest can be neglected. In fact, we have previously shown on the A2A adenosine G protein-coupled receptor (GPCR) that such an approach increases convergence and accuracy compared to larger systems [13] as long as local structural fluctuations of the active site are sufficiently sampled [26, 27].
To illustrate the applicability of QligFEP and assess the validity of the most commonly used parameters, as well as advising on how to change those, we tested our workflow on various systems previously used to benchmark other FEP protocols. This includes calculations of solvation free energies of side chain mimics [28], which represent a diverse chemical space in terms of size, polarity and atomic constitution. Thereafter, we apply this methodology to calculate relative binding free energies of 16 Cyclin-dependent kinase 2 (CDK2) ligands, and compare the performance of three different force field families (OPLS, CHARMM and AMBER) to previously published work [29]. Next, relative binding free energies of a series of antagonists for the adenosine A2A receptor are calculated and compared to a recently published approach [30, 31]. Finally, we show how the QligFEP dual-topology scheme can be applied for ‘scaffold hopping’, in the case of modifications of five Chk1 ligands of different chemotypes [32]. Our code and benchmark sets are available through GitHub (https://github.com/qusers/qligfep).
Materials and methods
Description of the workflow
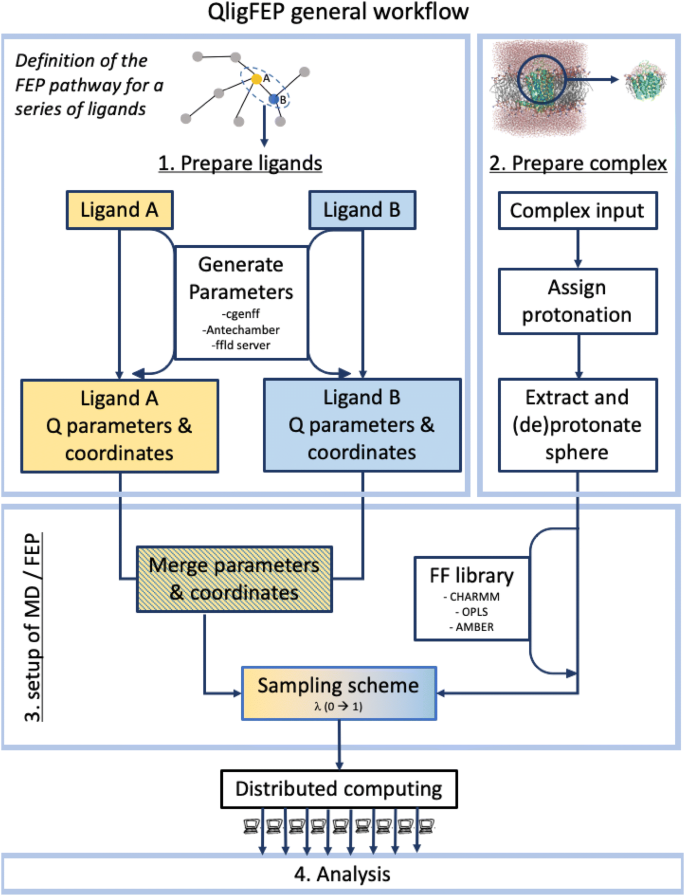
QligFEP is an application programming interface (API) that aims to automate and generalize the tedious process of setting up and analyzing the MD simulations for FEP calculations. Given a collection of molecules, for which we want to compare their relative binding affinities, a preliminary step involves the definition of the pair(s) of ligands to be compared. In QligFEP this can be done either manually, or by generating the pair list with an external program, e.g. LoMap [10], as represented in the top layer in Fig. 1. A good initial guess (adopted along this manuscript) is to define a radial pathway connecting all compounds to a central node (i.e. a single reference ligand). In cases where convergence is insufficient due to too big changes (assessed by a large SEM or even MD simulation crashes), one can add edges to nodes that involve smaller topological changes, as illustrated here in case of the A2AAR ligand binding calculations. The QligFEP workflow is then iteratively applied for each pair of molecules (A and B) in the pair list, and is composed of four modules (Fig. 1): (1) ligand parameterization, (2) complex preparation, (3) generation of the FEP and MD inputfiles and (4) an analysis tool.
General workflow of the QligFEP API, which consists of three preparation steps and one analysis module. The user only needs to provide .pdb coordinate files for the first two steps, from where parameter and library files are generated to perform the FEP calculations. Currently, the CHARMM, AMBER and OPLS forcefields are supported
Ligand parameterization
In the first module, the user provides PDB files of all small molecules that are going to be analyzed via FEP calculations. The coordinates of the molecules will be the input actually processed, i.e. this module does not enumerate protonation, enantiomeric and tautomeric states. The main aim of the module is thus to translate ligand 3D coordinates into Q readable library and parameter files, currently available for three families of force fields: OPLS-AA/M [33], Amber ff14sb [34] and CHARMM36 [3]. We provide a translation based on qtools [35] of the GAFF/AnteChamber [1, 36] parameters for AMBER. For CHARMM, the parameters are obtained and translated into Q format using CGenFF [37]. Our implementation of the OPLS force field for ligands includes the OPLS2005 version generated via Schrodinger’s ffld_server [38]. Note that one needs to obtain the required licenses for these external programs separately (which in some cases are free for academics). As an open source solution, LigParGen [5] can be used to generate Q readable parameter and library files based on the OPLS force field [5] and we are currently working to include other alternatives for ligand parameterization in the near future related e.g. to the OpenForceField consortium [39].
Complex preparation
In the second module, the macromolecular target is prepared for MD simulations of the binding site under spherical boundary conditions (SBC), as discussed in the introduction. The input is a PDB file, with or without hydrogens. In the former case the user should indicate which program has been used to add hydrogens, to account for the specific atom naming. If the module should, on the other hand, protonate the protein, the protonation state of ionizable residues is inferred from the residue name (e.g. HID, HIE, HIP for histidine protonated in delta, epsilon or positively charged, etc.), in accordance with the reference Q library files for proteins. This module also accounts for the two main parameters related to the implementation of the spherical boundary conditions in Q [40]: (1) the center of the sphere, which is normally placed at the center of geometry of a reference ligand, can be defined in different ways i.e. Cartesian coordinates, protein residue or a ligand atom, and (2) the size of the sphere, which should have a radius big enough such that it encompasses all the residues within the binding site, including a solvation patch to ensure sufficient dielectrical screening (e.g. 10–15 Å from the most distal atom in the ligand to the sphere boundary) [40]. The system is solvated with a pre-generated water grid in Q [23], and waters overlapping with either of the two ligands are automatically removed in the next stage. This module also accounts for the necessary neutralization of ionizable residues in the restrained area and outside the sphere dimensions [40].
Finally, this module also replicates the setup for the analogous calculations of the reference states for the FEP simulation, e.g. ligands in water for binding affinity calculations (or ligands in vacuum for the estimation of solvation free energies, see below).
Generation of the FEP and MD inputfiles
The third step prepares input files for the FEP simulations, according to a number of variables that should be carefully considered by the user. Given a pair of ligands A and B, their parameters (generated from step 1) are merged with the corresponding general force field parameters in Q, while the corresponding library files are merged into a dual topology library file (Fig. 1). These files are used together with the corresponding coordinates of the two ligands in the corresponding state, i.e. bound, solvated or vacuum, as indicated above, to build the topology file in each case. The subsequent FEP simulation, for which details are given below, will be guided by a number of input parameters, which are also generated at this stage and will define the desired sampling scheme. The FEP transformation involves sampling of a series of intermediate states along a pathway between the potentials, UA and UB, corresponding to each of the molecules. The probability density between the configurational distributions of the two states must overlap sufficiently. Therefore, the pathway is divided in a series of discrete steps following a linear combination of the potentials according to \(U_{i} = \left( {1 - \lambda_{i} } \right)U_{A} + \lambda_{i} U_{B}\), where the coupling parameter λt varies between 0 and 1, and an ensemble average is collected at each of these steps [41, 42]. It follows that the number of λ steps, their distribution along the FEP pathway (e.g. linear or non-linear), the sampling time per λ step, as well as the starting point (e.g., λ = 0, 0.5 or 1) and directionality of the simulations, are key parameters of the FEP simulation, which are defined in these modules. The simulations are setup as a number of replicate MD simulations (with different random initial velocities), and due to the convenience of using a distributed computing scheme (Fig. 1), we include the option to write out input files tailored for a specific (high performance) computing cluster.
Analyzing the simulations
The last module in QligFEP performs all required analysis to report free energies and the associated statistical parameters. This includes up to three different methods to calculate relative free energies: (1) Zwanzig’s exponential formula [43], (2) overlap sampling (OS) [44] and (3) Bennet’s acceptance ratio (BAR) [45], the latter method being the one reported throughout this manuscript. Standard errors of the mean (SEM) are estimated from the individual replicate simulations, The module provides a linear-regression statistical analysis (i.e. for retrospective analysis when experimental affinity values are available) including the Pearson R2 with associated 95% confidence intervals (CI, p = 0.05, based on the application of Fisher’s z transformation for application to small datasets [46]), and the mean absolute error (MAE), calculated using scikit-learn [47]. Further details as to the specifications of the command line input can be found in the manual, which includes several tutorials.
System preparation
Prior to entering the workflow, protein and ligand coordinates must be generated and/or retrieved from the corresponding databases. We herein describe the details of this procedure for the dataset treated in this manuscript.
Ligand coordinates were generated in a reasonable 3D conformation with the appropriate tools in Maestro [48]. The starting point differed between the different test cases, i.e., drawing from scratch (the side-chain mimics), modifying the crystal coordinates of the reference ligand (scaffold hopping) or the docked poses kindly provided by Wang et al. (CDK2 inhibitors) [49], or those obtained by manual docking (A2AAR antagonists). Each molecule was subsequently treated as the input for the corresponding parameterization tools, managed by the “Ligand preparation” module as described above.
All protein structures were pre-processed using Protein Preperation Wizard in Maestro [50]. In the case of the A2AAR, the receptor was embedded in a lipid bilayer and shortly equilibrated using the protocols in the GPCR-ModSim web server (http://www.gpcr-modsim.org) as described elsewhere [13, 51,52,53]. Selected water molecules in the binding site (from the crystal structures of CDK2 and Chk1, or from the or the MD equilibration in the case of A2AAR) were initially retained, and the complex given as input for the “Complex preparation” module. In each case, the center of the sphere was automatically generated from the center of geometry of the two ligands involved in the perturbation, and the sphere radius was initially set to 25 Å, which is a good guess in most cases considering the typical size of drug-like molecules [40]. However, in the case of CDK2 inhibitors the radius was optimized to 22 Å as a combination of two factors: the relatively small size of the ligands in the dataset, and the fact that this smaller radius allowed to exclude from the simulated sphere a highly charged cluster of residues formed by Arg50, Arg126, Arg150 and a phosphorylated Thr160, which would otherwise not have enough solvation patch as they would lie on the sphere boundary [40].
MD/FEP simulations
The software package Q [23] was used for the MD sampling under SBC with the following settings: atoms lying outside the simulation sphere were tightly constrained to their initial coordinates (200 kcal/mol/Å2 force constant) and excluded from the calculation of the non-bonded interactions. In the boundary of the sphere, solvent atoms subject to polarization and radial restrains using the surface constrained all-atom solvent (SCAAS) [23, 54] model to mimic the properties of bulk water at the sphere surface. Long range electrostatics interactions beyond a 10 Å cut off were treated with the local reaction field method [55], except for the atoms undergoing the FEP transformation where no cutoff was applied. Solvent bonds and angles were constrained using the SHAKE algorithm [56].
The system was then subjected to 10 parallel MD replicate simulations, which only differed in their initially assigned random (Maxwell-Boltzman) velocities. Each simulation includes an equilibration scheme as follows, which is the default in QligFEP: (1) an initial phase of 31 ps where the simulation sphere is heated from 0.1 to 298 K and a positional restraint of 25 kcal/mol/Å2 initially imposed on all solute heavy atoms slowly released; (2) a 100 ps unbiased and unrestrained equilibration. The subsequent production phase followed, where the sampling along a λ transformation pathway (defining the FEP transformation) was performed with the following parameters in the different datasets (unless indicated the contrary): the transformation was divided into 51 λ steps, evenly distributed (i.e. linear sampling), the corresponding MD sampling for each λ step being 10 ps using a 1 fs time step. Thus, each FEP transformation involved a total simulation time of 10 × (131 ps + (51 × 10 ps)) = 6.41 ns for each of the two legs in the thermodynamic cycle, resulting in a total sampling time of 12.82 ns per perturbation. For comparison, a typical perturbation in the commercial alternative FEP+ includes one simulation of 12 lambda windows, with 5 ns sampling per window giving 120 ns under a bigger system using PBC [30]. However, in cases where large changes are considered (i.e. more than 6 heavy atoms) the sampling above might be insufficient to achieve the desired convergence, which might be easily diagnosed by a high SEM or even simulation crashes. In such cases, one can increase the phase-space overlap by increasing the λ windows, or either using a more dense (non-linear) sampling around the initial/ending states, or just increase the sampling of each λ window to increase convergence. In this work, a different scheme was used for the larger perturbations involved in the datasets of the relative hydration free energies and the A2AAR antagonist binding.
In the dual topology paradigm, each perturbation involves parallel growing (A) or annihilation (B) of the relative atoms of the two ligands A and B, and includes the perturbation to/from soft-core potentials as an intermediate step to ensure sufficient convergence [57]. Half-harmonic distance restraints of 2.0 kcal/mol/Å2 were applied to maintain pairs of equivalent (non-dummy) atoms between the two ligands (A and B), within a window distance of 0.0–0.2 Å. Since this restraint is only applied on atom pairs, of which one of them will have the interactions fully turned off in each of the end states, the energetic term of the restraints cancels and no additional correction needs to be applied. The relative free energy (∆∆GA-B) is calculated by closing the corresponding thermodynamic cycle, where the ∆G corresponding to each of the two legs was calculated with the BAR approach [45]. When the ligand mutation involves a change in the total charge of the sphere, a Born correction term was added to the calculated free energies to account for the polarization effect [58], estimated as:
where QI is the net charge of the solute, and rBorn is the radius of the cavity in the macroscopic medium, with a dielectric constant ε, in which the charge is embedded.
All calculations were performed using the OPLS-AA/M [33] force field with TIP3P waters, except for the CDK2 inhibitor set where results are reported with the three families of force fields implemented in QligFEP. Standard errors of the mean (SEM) are estimated from the individual replicate simulations by QligFEP, whereas errors reported for FEP+ are either BAR analytical error estimations or bootstrap estimated errors as reported in their original publications [49].
Results and discussion
In this section, we use the workflow described above in four different case scenarios, where the calculation of relative free energies can be correlated with experimental hydration or binding free energies for different series of molecules as reported in the literature. The first case accounts for estimation of solvation free energies, tested on the Wolfenden set of protein side chain mimics [28]. Next, QligFEP is used to automatically estimate relative binding affinities of ligand series in two different target systems: (1) a globular enzyme, cyclin-dependent kinase 2-cyclin A receptor (CDK2), where relative affinities of a dataset of 16 inhibitors are also used to evaluate the accuracy of the three different force fields implemented, and (2) antagonist binding to a G-protein coupled receptor, the adenosine receptor A2A (A2AAR). Finally, we show how our perturbation protocol can accurately assist ‘scaffold hopping’ [59] modifications on a series of five Checkpoint kinase 1 (Chk1) inhibitors.
Relative hydration free energies
The solubility of small molecules is a key physicochemical property of any drug candidate [60]. Therefore, there is much interest in accurate computational estimations of ligand solubility via e.g. calculation of their corresponding solvation free energies. FEP has been quite successfully applied to obtain such estimations, although force field and water models can significantly influence accuracy [61]. To calculate the water solvation (hydration) free energies, a thermodynamic cycle can be constructed where the calculation legs refer to perturbations of the molecules of interest in vacuum and water, respectively. Our test case consists of mimics of amino acid side chains, for which absolute hydration free energies have been well characterized [28]. Notably, these experimental values were determined in a way that, in case of ionizable residues, only neutral states of the side chain mimic were considered. Thus, to be able to compare the solvation free energies of the corresponding charged species, the hydration free energy of the proton needs to be taken into account [62]. Here we use the values reported by Tissandier et al. [63], as adopted by Kelly et al. [64] and included in the Minnesota solvation database [65, 66]. For a more detailed discussion on the hydration free energy of the proton we refer to the work of Zhang et al. [62]. The total set thus includes 23 amino acid mimics (Table 1), encompassing diverse molecules in terms of physicochemical properties for which hydration free energies cover an energetic span of more than 80 kcal/mol. Taking the smallest methyl group sidechain in alanine as a reference, we calculated relative hydration free energies between any given side chain mimic (X) to methane (Me), and compared to the relative hydration free energies provided in Wolfenden et al. (note that, given this strategy, glycine was excluded) [28]. The dual topology framework allows us to perform the calculations in three alternative ways: sidechain annihilation (X → Me), sidechain growing (Me → X), both following a classical λ (1 → 0) FEP pathway, or alternatively starting from a mixture of both states (λ = 0.5) and propagating in each direction (1 ← λ → 0). As reported in Table 1, the simulations are well converged with a very low SEM (± 0.23 kcal/mol on average) over all replicate simulations in each of the three cases. The hysteresis, defined as the difference between the values obtained in the forward (annihilation) and backward (creation) directions, is also generally very low, with an average value of 0.47 kcal/mol. Notably, in the cases involving the largest growth (i.e. for p-cresol and 3-methylindole in the Me → X pathway), the default sampling with 51 λ windows was not sufficient to ensure convergence, as none of the simulations ran to completion. In these cases, a second round with 101 λ windows were needed and the corresponding values reported in Table 1. This is a first example indicating that low convergence may be a criterion for revising the FEP strategy (see below for A2A antagonists). Regardless of the perturbation scheme applied, the calculated solvation free energies show an overall excellent agreement with experiment, with an almost perfect correlation in terms of R2. Given the large span of free energies, this value indeed masks the accumulated deviation from experiment, and the R2 determined on the separate charged/neutral groups is around 0.9 for all methods (see Table 1). While the calculated MAE over the whole dataset falls within a range of 1.46–1.76 kcal/mol, most of this error is caused by the higher deviations for ionizable molecules, with a smaller error below 1 kcal/mol in the best case for neutral sidechains. This is to be expected, as it is well known that the accuracy of force fields for these moieties is lower, and various corrections are needed to account for artifacts introduced by periodicity and long-range electrostatic cut-offs [62]. In our case, the latter does not apply since simulations are performed in a finite spherical droplet of water, where no cut-off is used for any atoms undergoing the alchemical transformation, allowing us to apply a simple correction to the calculated free energies of charged species using Born’s formula [58]. This yields an overall MAE of 3.67 kcal/mol for the five charged residues, which is considerably lower than those reported with the same force field by Zhang et al. [62], which range from 5.52 to 20.23 kcal/mol depending on the correction applied. It is also interesting to compare the results of QligFEP to other computational studies which, using a variety of sampling techniques and force fields, did not report any calculations on the charged species [67,68,69,70]. The MAE obtained in the study considering the neutral form of the aminoacid mimics is 1.71 kcal/mol, which is outperformed by our results (Table 1, considering only non-charged residues MAE = 0.95–1.24 kcal/mol). Indeed the lowest MAE (0.69 kcal/mol) was reported from the Sandler group excluding any form of Asp, Glu, Lys or Arg [67], which in our case would yield an even lower value of MAE = 0.53 kcal/mol.
While the complete annihilation of a sidechain seems to work well in all cases, we noted that the sidechain growth needs a smoother protocol to be implemented for bigger substituents (e.g. see Me to p-cresol or 3-methylindole). Thus, we have adopted the ‘middle’ scheme (1 ← λ = 0.5 → 0) as the standard protocol throughout the rest of this manuscript, since it forms the best trade-off between general applicability and accuracy. There are cases where the user might want to have more control over endpoints, introduce a smoother lambda spacing scheme, or calculate averages over two endpoint simulations, which is easily achieved using QligFEP.
Ligand binding to CDK2
The first example of ligand binding involves a series of antagonists of the CDK2 receptor [71]. This receptor is a drug target in oncology, and several inhibitors have been identified. Indeed, many solved CDK2—inhibitor crystal structures exist, making this system particularly suitable for testing structure-based computational methods. In an earlier FEP study on this system, Wang et al. [49] examined the relative affinities for a set of 16 inhibitors using FEP+, which included 6 crystallized inhibitor-protein complexes and 10 analogs of these inhibitors, with a manually designed cycle closure and the OPLS2005 force field. Later on, the same dataset was used to test the automated FEP workflow implemented in FEP+, in combination with the new proprietary versions of the OPLS force field [17, 29]. We herein use this dataset to evaluate the performance of QligFEP in a (retrospective) ligand design project. Figure 2 shows the binding mode of the main scaffold (panel A) and the perturbation scheme applied in this study (panel B). The six structures solved in complex with CDK2 are depicted as square nodes, with their PDB codes indicated, while the remainder of the ligands (ellipsoidal boxes) were considered in the docking poses kindly provided by Wang et al. All substituents involved different modifications of the benzene ring of ligand 1h1q, which forms the central node in our FEP strategy. The results are summarized in Table 2 and Fig. 3, which also includes the values obtained with Schrodinger’s FEP+ for comparative purposes [29], together with the relevant statistical figures of merit. The affinity values, computed relative to the reference compound 1h1q, are here transformed to absolute binding free energies by scaling to the affinity of this compound, to facilitate the comparison with the FEP+ results. QligFEP results show very good agreement with the experimental data, in particular with the OPLS force field. The low MAE obtained (0.85 kcal/mol) is very encouraging and only slightly higher than the MAE computed from the FEP+ results. The correlation of QligFEP-OPLS results with experimental data, on the other hand, is the highest of the methods compared. Not only does this model result in the best correlation coefficient, but the statistics actually denote an important improvement in the predictive power and interpretation of this model as compared to all other models in Table 2, with a slope very close to the ideal value of 1 (Fig. 3) and the smallest intercept of 1 kcal/mol. These values are in contrast with the two FEP+ models and the QligFEP models obtained with the AMBER and CHARMM force fields, where the slope is around 0.5 and the intercept close to − 5 kcal/mol, indicating that most of the variability of the data is actually not explained by the models (Fig. 3). We should stress that in QligFEP no attempts have been made to increase the quality of the ligand parameters generated by any of the automated parameterization methods. In particular, when using either GAFF or CGenFF, quite a few critical warnings were ignored, which in a real (prospective) application one would certainly optimize, and this could partially account for the lower performance as compared to the results using the OPLS force field. However, we feel that such an optimization procedure would be beyond the scope of this work, as the aim of the reported tool is to automate FEP calculations, not parameter generation, and the possibility of using different force fields as per the preference (and experience) of the user. In this sense, we would also like to stress that the QligFEP calculations were performed using the OPLS-AA/M model, which is very similar to OPLS2005 but supposedly inferior to the latest version OPLS3, a proprietary version not publicly available. However, as stated before the performance of OPLS3 with a more complex cycle closure strategy only improves slightly in terms of the MAE (0.1 kcal/mol), whilst the R2 and interpretation of the model is superior in QligFEP.
a Binding mode of CDK2 inhibitor 17, showing key interactions with the backbone in the hinge region of the protein. All substituents introduced at R are positioned in a solvent exposed cavity on the surface on the protein. b Overview of the chemical constitution of the 16 R-groups (nodes) and selected perturbations (edges) for the calculations reported in Table 2
Scatter plots for the calculated and experimental relative binding free energies (∆Gbind, kcal/mol) for the series of 16 CDK2 inhibitors, taking 1h1r as reference. The orange line represents the calculated linear equation for the correlation coefficient R2. The black line represents a perfect correlation, and the two dashed lines are +/− 1 and +/− 2 kcal/mol respectively
A2AAR antagonist binding
The A2AAR has been widely used by us and others as a test-case for the performance of computational methods in the design of ligands for more complex membrane systems [72]. The advantage of using spherical boundary conditions becomes evident in these systems, where a sphere centered on the binding site with a diameter of 50 Å contains approximately 7.400 atoms (see Fig. 1). This significantly decreases the computational time needed for a simulation as compared to a periodic system of i.e. an optimized hexagonal shaped box with ~ 42.000 atoms. We have shown that calculations performed on larger sphere systems of the same A2AAR receptor complex result in larger errors, but yield the same average changes in binding free energies [12, 73]. On the related A3AR, our FEP approach now automated under QligFEP was used to elucidate the role of a nitrogen substitution in the core of antagonists with a pyrimidine scaffold [74]. Here, we apply QligFEP to compute the relative affinities of 8 analogs of the triazol-2-yl-9 H-purin-6-ylamine (ST1535, Fig. 4) [31]. The binding mode of this compound, shown in Fig. 4, was inferred by analogy to the binding mode of the triazolopyrimidine antagonist ZM241385 [75]. This includes key hydrogen bonds with N2536.55 and E169EL2 (subscripts indicate the general GPCR topological nomenclature [31]) and π–π stacking interactions between the bicyclic core of the ligand and F168EL2, which are common interactions in adenosine receptor ligand recognition [76]. Table 3 reports the calculated relative free energy (∆∆Gbind) of each compound to the simplest ligand compound 11. For comparison, the experimental values are shown together with the values obtained by Lenselink et al. [30] using FEP+ under PBC. In a first attempt, each of the eight analogs was directly perturbed to the reference ligand 11 (note the nomenclature of the compounds following the original report by Minetti et al. [31]). Six out of the eight perturbations are well converged, whereas we observed a large SEM (over 1.6 kcal/mol) for the perturbations including the largest compounds 25c and 25d. This provides some insight in the convergence limitations of a direct perturbation between two related ligands. In this case, it was evidenced that the phenylethyl (25d) or pentyl (25c) substitutions are too large to compute reliable relative affinities when directly perturbed to hydrogen (11). To solve this, we created an alternative pathway that involved a smoother annihilation scheme through two intermediate steps linking these ligands to the reference compound 11, i.e. 25d → 25c → 25b → 11. The calculated relative affinities with QligFEP show excellent agreement with the experimental data (Table 3). Moreover, the statistical figures of merit (R2 = 0.88; MAE = 0.72 kcal/mol) are equivalent to those reported by Lenselink et al. (R2 = 0.78; MAE = 0.68 kcal/mol). Notably, in that work the proprietary OPLS3 force field was used and the level of performance indicated was only achieved after applying the cycle-closure strategy, which includes a total of 17 perturbations in redundant cycles and the transformation of the data to include ∆G values relative to 11. If only the perturbations to 11 are selected and the relative binding free energies are used (which give a shorter span and thereafter are more sensible to fluctuations in the overall correlation), the correlation of the FEP+ strategy is moderately hampered (R2 = 0.50, as opposed to the corresponding value of R2 = 0.74 for QligFEP). It is worth noting that two perturbations that we accurately model include changes in the core ring (32 and 41–11), which belong to the scaffold hopping modifications that we discuss in more detail below.
Scaffold hopping
Thus far, examples presented in this work involved typical lead-optimization strategies, where R-group substituents are introduced to a scaffold with the aim to increase binding affinity. Another strategy within the medicinal chemist’s toolbox is to change the core of the lead ligand while retaining peripheral R-group substituents that are identified as crucial for protein–ligand recognition [59, 77]. Such a procedure, also referred to as ‘scaffold-hopping’, can be extremely useful to overcome problems related to the initial scaffold, such as ADMET properties or reactivity, as well as to expand the chemical space and overcome patentability problems [77]. However, scaffold hopping has been elusive to FEP approaches as it often involves significant amounts of changes in the bond topologies within the ligand series. Within a single topology framework, one would have to make or break bonds to include such changes, something that is prone to numerical instabilities due to the asymptotic nature of harmonic potentials representing bond stretching/closing in classical force fields [21, 78]. One strategy to overcome this is to change the asymptotic nature of the bond by introducing a softcore bond potential [78]. Within the QligFEP dual-topology framework, we introduce an alternative approach, since no bonded terms are altered within the transition of the ligand pair, even if the bond topology changes. Additionally, since the two molecules exist as two unrelated entities, the necessary dummy atoms do not experience any strain from residual ‘real’ atoms, and the thermodynamic cycle can be formally closed [21]. To test the applicability of this approach in a typical scaffold-hopping case, we performed a total of six perturbations between five inhibitors of the Checkpoint kinase 1 (Chk1, see Fig. 5). This set was extracted from a recent benchmark on scaffold hopping using FEP+ applying the aforementioned softcore potentials, and it is illustrative of the various topological changes observed in these type of modifications, i.e. ring opening, ring closure, changing the ring size and the substitution of atoms within a ring. The results are summarized in Table 4. Even if the changes in affinity within the series are relatively small (which is typically the case in a successful scaffold hoping project, i.e. to discover new chemical entities with similar binding affinities) these are very successfully captured with our QligFEP approach, with an MAE of 0.32 kcal/mol. More importantly, with the only exception of the pair 19 → 21 involving a ring formation, see Fig. 5, the simulations are well converged with a SEM typically under 0.5 kcal/mol (Table 4). Notably, these values are quite comparable with those reported with the alternative FEP+ approach [78], even though a different force field version was used (OPLS2005 in our methods versus OPLS3 in FEP+, see Table 4). This shows that the practical advantages of a dual topology approach in scaffold hopping do not cause a lower performance as compared to single topology approaches.
Core of the scaffold of the Chk1 inhibitor series, showing the variable position explored by scaffold hopping with the five different R-group modifications (bottom). The pathways chosen to connect these groups through FEP simulations to calculate the relative change in binding free energies are indicated by arrows, the corresponding values reported in Table 4
Discussion
We present QligFEP, an automated workflow to setup, run and analyze ligand free energy perturbation calculations within a structure-based drug design framework. The workflow is built as an API that interacts with the open-source software package Q. It aims to automate the tedious process of creating input and parameters files and to facilitate a robust setup to perform free energy calculations between a given pair of ligands to calculate e.g. their relative binding affinities. We show how this approach can be scaled up to perform pair comparisons in a high-throughput fashion for a versatile set of ligands and receptors, which to the best of our knowledge is achievable with a limited amount of software packages [9, 29, 49].
Given a dataset with a number l of ligands, one can eventually perform all possible pair comparisons, which is proportional to l2. This brings up the question on how to optimize the design of the FEP strategy such that a selected sample of pair comparisons (li, lj) represent the spectrum of relative affinities to cover the whole dataset whilst minimizing the structural changes involved in order to ensure sufficient convergence. One way to approach this problem is through the use of MCS algorithms, further combined with the idea of cycle closures, to validate the FEP pathways (i.e., the addition to the ∆∆Gbind calculated along a given path A → B → C → A should be zero). While for big datasets this automated approach is a useful solution, in most cases one can design pathways based on basic medicinal chemistry knowledge, and assess the feasibility of this design solely based on the convergence achieved. This is nicely illustrated in this work by the calculations performed on the A2AAR antagonists set. The initial design of the FEP pathway involved the calculation of the affinities of each compound relative to a single reference compound. This is, in our opinion, the recommended approach in a lead optimization project, where typically one only knows the affinity of a reference compound and variations of one or several substituents are designed. In this case, two perturbations showed low convergence, detected by an abnormally high SEM within the replicate simulations of the system. We redesigned the strategy to circumvent this issue, involving a cycle closure defined by four compounds within the dataset. This case also illustrates the increased computational efficiency of QligFEP as compared to the FEP+ methodology. First, the cycle-closure strategy used in FEP+ involved 17 perturbations as compared to the 10 perturbations in our FEP pathway. Second, our system setup with SBC is approximately 6 times smaller than the PBC setup used by FEP+. Third, the simulation times reported are 10 times longer (see methods section) for a single simulation per ligand pair. Taken all together, QligFEP provides an alternative for efficient FEP simulations, at a substantial reduction of the computational cost as compared with existing commercial software. In other words, we show an alternative to a more exhaustive cycle closure, which is based on starting with a simple FEP design based on the MCS idea, detect potential problematic pairs based on convergence using the SEM as a metric and iterative processing of the obtained data if needed.
Another advantage of our approach is the implementation of a dual topology representation, which allows the comparison of molecules with unrelated topologies. This is a particularly interesting approach in, for instance, scaffold hopping where one wants to normally compare different chemical scaffolds. In a single topology framework, where a one-to-one mapping of atoms between the two states is used, this potentially includes the transformation of bonded terms to represent the change in bond topology. In contrast, with QligFEP this problem can be circumvented, whilst still achieving excellent correlation with experiment. Our initial results are promising, and the next direction in our research will be to apply the QligFEP dual topology approach the binding affinity prediction of fragment-like compounds, which involves comparing different chemical scaffolds where even potential multiple binding modes can be assessed.
Conclusion
QligFEP provides a versatile, robust and accurate framework to routinely perform FEP calculations in structure-based ligand design projects. We herein show the performance on a diverse benchmark set of systems and ligands, showing agreement between experiment and calculations similar to that of other state-of-the-art methods. QligFEP is implemented as an easy to use, modular API, which interacts with the open-source MD engine Q. Additionally, our package already works with a number of standard force fields, whilst the implementation of additional force fields is currently under development. The benchmark test sets included in this study are part of the tutorials and provide a useful start for users with limited training in free energy calculations. QligFEP and its associate MD engine Q are available free of charge through github.
Abbreviations
- BAR:
-
Bennet’s acceptance ratio
- CDK2:
-
Cyclin-dependent Kinase 2
- FEP:
-
free energy perturbation
- GPCR:
-
G protein-coupled receptor
- MAE:
-
mean unassigned error
- MD:
-
molecular dynamics
- PBC:
-
periodic boundary conditions
- SBC:
-
spherical boundary conditions
- SEM:
-
standard error of the mean
References
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general Amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Hornak V, Abel R, Okur A et al (2006) Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Genet 65:712–725
Huang J, Mackerell AD (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34:2135–2145. https://doi.org/10.1002/jcc.23354
Robertson MJ, Tirado-Rives J, Jorgensen WL (2015) Improved peptide and protein torsional energetics with the OPLS-AA force field. J Chem Theory Comput 11:3499–3509. https://doi.org/10.1021/acs.jctc.5b00356
Dodda LS, De Vaca IC, Tirado-Rives J, Jorgensen WL (2017) LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res 45:W331–W336. https://doi.org/10.1093/nar/gkx312
Il Lee K, Rui H, Pastor RW, Im W (2011) Brownian dynamics simulations of ion transport through the VDAC. Biophys J 100:611–619. https://doi.org/10.1016/j.bpj.2010.12.3708
Tembre BL, Mc Cammon JA (1984) Ligand-receptor interactions. Comput Chem 8:281–283. https://doi.org/10.1016/0097-8485(84)85020-2
Cournia Z, Allen B, Sherman W (2017) Relative binding free energy calculations in drug discovery: recent advances and practical considerations. J Chem Inf Model 57:2911–2937
Loeffler HH, Michel J, Woods C (2015) FESetup: automating setup for alchemical free energy simulations. J Chem Inf Model 55:2485–2490. https://doi.org/10.1021/acs.jcim.5b00368
Liu S, Wu Y, Lin T et al (2013) Lead optimization mapper: automating free energy calculations for lead optimization. J Comput Aided Mol Des 27:755–770. https://doi.org/10.1007/s10822-013-9678-y
Frenkel D, Smit B, Ratner MA (2008) Understanding molecular simulation: from algorithms to applications. Phys Today 50:66. https://doi.org/10.1063/1.881812
Boukharta L, Gutiérrez-de-Terán H, Åqvist J (2014) Computational prediction of alanine scanning and ligand binding energetics in G-protein coupled receptors. PLoS Comput Biol 10:e1003585. https://doi.org/10.1371/journal.pcbi.1003585
Keränen H, Åqvist J, Gutiérrez-de-Terán H (2015) Free energy calculations of A 2A adenosine receptor mutation effects on agonist binding. Chem Commun 51:3522–3525. https://doi.org/10.1039/C4CC09517K
Gapsys V, Michielssens S, Seeliger D, de Groot BL (2015) pmx: automated protein structure and topology generation for alchemical perturbations. J Comput Chem 36:348–354. https://doi.org/10.1002/jcc.23804
Polishchuk PG, Madzhidov TI, Varnek A (2013) Estimation of the size of drug-like chemical space based on GDB-17 data. J Comput Aided Mol Des 27:675–679. https://doi.org/10.1007/s10822-013-9672-4
Reymond JL (2015) The chemical space project. Acc Chem Res 48:722–730. https://doi.org/10.1021/ar500432k
Wang L, Wu Y, Deng Y et al (2015) Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J Am Chem Soc 137:2695–2703. https://doi.org/10.1021/ja512751q
Homeyer N, Gohlke H (2013) FEW: a workflow tool for free energy calculations of ligand binding. J Comput Chem 34:965–973. https://doi.org/10.1002/jcc.23218
Christ CD, Fox T (2014) Accuracy assessment and automation of free energy calculations for drug design. J Chem Inf Model 54:108–120. https://doi.org/10.1021/ci4004199
Michel J, Essex JW (2010) Prediction of protein-ligand binding affinity by free energy simulations: assumptions, pitfalls and expectations. J Comput Aided Mol Des 24:639–658
Liu S, Wang L, Mobley DL (2015) Is ring breaking feasible in relative binding free energy calculations? J Chem Inf Model 55:727–735. https://doi.org/10.1021/acs.jcim.5b00057
Gapsys V, Michielssens S, Seeliger D, De Groot BL (2015) pmx: automated protein structure and topology generation for alchemical perturbations. J Comput Chem 36:348–354. https://doi.org/10.1002/jcc.23804
Marelius J, Kolmodin K, Feierberg I et al (1998) Q: a molecular dynamics program for free energy calculations and empirical valence bond simulations in biomolecular systems. J Mol Graph Model 16:213–225. https://doi.org/10.1016/S1093-3263(98)80006-5
Bauer P, Barrozo A, Purg M et al (2018) Q6: a comprehensive toolkit for empirical valence bond and related free energy calculations. SoftwareX
Lee F, Warshel A (1992) A local reaction field method for fast evaluation of long-range electrostatic interactions in molecular simulations. J Chem Phys 97:3100–3107
Isaksen GV, Åqvist J, Brandsdal BO (2016) Enzyme surface rigidity tunes the temperature dependence of catalytic rates. Proc Natl Acad Sci 113:7822–7827. https://doi.org/10.1039/c7cc00980a
Bjelic S, Brandsdal BO, Åqvist J (2008) Cold adaptation of enzyme reaction rates. Biochemistry 47:10049–10057. https://doi.org/10.1021/bi801177k
Wolfenden R, Andersson L, Cullis PM, Southgate CCB (1981) Affinities of amino acid side chains for solvent water. Biochemistry 20:849–855. https://doi.org/10.1021/bi00507a030
Harder E, Damm W, Maple J et al (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296. https://doi.org/10.1021/acs.jctc.5b00864
Lenselink EB, Louvel J, Forti AF et al (2016) Predicting binding affinities for GPCR ligands using free-energy perturbation. ACS Omega 1:293–304. https://doi.org/10.1021/acsomega.6b00086
Minetti P, Tinti MO, Carminati P et al (2005) 2-n-butyl-9-methyl-8-[1–3]triazol-2-yl-9H-purin-6-ylamine and analogues as A2A adenosine receptor antagonists. Design, synthesis, and pharmacological characterization. J Med Chem 48:6887–6896. https://doi.org/10.1021/jm058018d
Wang L, Deng Y, Wu Y et al (2017) Accurate modeling of scaffold hopping transformations in drug discovery. J Chem Theory Comput 13:42–54. https://doi.org/10.1021/acs.jctc.6b00991
Robertson MJ, Tirado-Rives J, Jorgensen WL (2015) Improved peptide and protein torsional energetics with the OPLS-AA force field. J Chem Theory Comput 11:3499–3509. https://doi.org/10.1021/acs.jctc.5b00356
Maier JA, Martinez C, Kasavajhala K et al (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11:3696–3713. https://doi.org/10.1021/acs.jctc.5b00255
Purg M, Bauer P (2017) qtools v0.5.9
Wang J, Wang W, Kollman PA, Case DA (2001) Antechamber, an accessory software package for molecular mechanical calculations. J Am Chem Soc 222:U403. https://doi.org/10.1016/j.jmgm.2005.12.005
Vanommeslaeghe K, Hatcher E, Acharya C et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. https://doi.org/10.1002/jcc.21367
Banks JL, Beard HS, Cao Y et al (2005) Integrated modeling program, applied chemical theory (IMPACT). J Comput Chem 26:1752–1780
Mobley DL, Bannan CC, Rizzi A et al (2018) Open Force Field Consortium: Escaping atom types using direct chemical perception with SMIRNOFF v0.1. bioRxiv 1–36. https://doi.org/10.1101/286542
Gutiérrez-De-Terán H, Åqvist J (2012) Linear interaction energy: method and applications in drug design. Methods Mol Biol 819:305–323. https://doi.org/10.1007/978-1-61779-465-0_20
Pohorille A, Jarzynski C, Chipot C (2010) Good practices in free-energy calculations. J Phys Chem B 114:10235–10253. https://doi.org/10.1021/jp102971x
Brandsdal BO, Österberg F, Almlöf M et al (2003) Free energy calculations and ligand binding. Adv Protein Chem 66:123–158
Zwanzig RW (1954) High-temperature equation of state by a perturbation method. I. Nonpolar gases. J Chem Phys 22:1420. https://doi.org/10.1063/1.1740409
Kirkwood JG (1935) Statistical mechanics of fluid mixtures. J Chem Phys 3:300–313. https://doi.org/10.1063/1.1749657
Bennett CH (1976) Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys 22:245–268. https://doi.org/10.1016/0021-9991(76)90078-4
Fisher RA (1921) On the “probable error” of a coefficient of correlation deduced from a small sample. Metron 1:3–32
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830
Schrödinger Release 2015-4
Wang L, Deng Y, Knight JL et al (2013) Modeling local structural rearrangements using FEP/REST: application to relative binding affinity predictions of CDK2 inhibitors. J Chem Theory Comput 9:1282–1293. https://doi.org/10.1021/ct300911a
Madhavi Sastry G, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Keränen H, Gutiérrez-de-Terán H, Åqvist J (2014) Structural and energetic effects of A2A adenosine receptor mutations on agonist and antagonist binding. PLoS ONE 9:e108492. https://doi.org/10.1371/journal.pone.0108492
Esguerra M, Siretskiy A, Bello X et al (2016) GPCR-ModSim: a comprehensive web based solution for modeling G-protein coupled receptors. Nucleic Acids Res 44:W455–W462. https://doi.org/10.1093/nar/gkw403
Gutiérrez-de-Terán H, Bello X, Rodríguez D (2013) Characterization of the dynamic events of GPCRs by automated computational simulations. Biochem Soc Trans 41:205–212. https://doi.org/10.1042/BST20120287
King G, Warshel A (1989) A surface constrained all-atom solvent model for effective simulations of polar solutions. J Chem Phys 91:3647. https://doi.org/10.1063/1.456845
Lee FS, Warshel A (1992) A local reaction field method for fast evaluation of long-range electrostatic interactions in molecular simulations. J Chem Phys 97:3100. https://doi.org/10.1063/1.462997
Ryckaert J-PJ, Ciccotti G, Berendsen HJH (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341. https://doi.org/10.1016/0021-9991(77)90098-5
Beutler TC, Mark AE, van Schaik RC et al (1994) Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chem Phys Lett 222:529–539. https://doi.org/10.1016/0009-2614(94)00397-1
Åqvist J (1990) Ion-water interaction potentials derived from free energy perturbation simulations. J Phys Chem 94:8021–8024. https://doi.org/10.1021/j100384a009
Zhao H (2007) Scaffold selection and scaffold hopping in lead generation: a medicinal chemistry perspective. Drug Discov Today 12:149–155
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and developmental settings. Adv Drug Deliv Rev 23:3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Chodera JD, Mobley DL, Shirts MR et al (2011) Alchemical free energy methods for drug discovery: progress and challenges. Curr Opin Struct Biol 21:150–160
Zhang H, Jiang Y, Yan H et al (2017) Free-energy calculations of ionic hydration consistent with the experimental hydration free energy of the proton. J Phys Chem Lett 8:2705–2712. https://doi.org/10.1021/acs.jpclett.7b01125
Tissandier MD, Cowen KA, Feng WY et al (1998) The proton’s absolute aqueous enthalpy and Gibbs free energy of solvation from cluster-ion solvation data. J Phys Chem A 102:7787–7794. https://doi.org/10.1021/jp982638r
Kelly CP, Cramer CJ, Truhlar DG (2006) Aqueous solvation free energies of ions and ion-water clusters based on an accurate value for the absolute aqueous solvation free energy of the proton. J Phys Chem B 110:16066–16081. https://doi.org/10.1021/jp063552y
Marenich A, Kelly C, Thompson J, Hawkins G (2012) Minnesota solvation database. Minnesota Solvation Database version 20
Palascak MW, Shields GC (2004) Accurate experimental values for the free energies of hydration of H+, OH−, and H3O+. J Phys Chem A 108:3692–3694. https://doi.org/10.1021/jp049914o
Chang J, Lenhoff AM, Sandler SI (2007) Solvation free energy of amino acids and side-chain analogues. J Phys Chem B 111:2098–2106. https://doi.org/10.1021/jp0620163
Shirts MR, Pitera JW, Swope WC, Pande VS (2003) Extremely precise free energy calculations of amino acid side chain analogs: comparison of common molecular mechanics force fields for proteins. J Chem Phys 119:5740–5761. https://doi.org/10.1063/1.1587119
Maccallum JL, Peter Tieleman D (2003) Calculation of the water-cyclohexane transfer free energies of neutral amino acid side-chain analogs using the OPLS all-atom force field. J Comput Chem 24:1930–1935. https://doi.org/10.1002/jcc.10328
Duarte Ramos Matos G, Kyu DY, Loeffler HH et al (2017) Approaches for calculating solvation free energies and enthalpies demonstrated with an update of the FreeSolv database. J Chem Eng Data 62:1559–1569
Hardcastle IR, Arris CE, Bentley J et al (2004) N2-substituted O6-cyclohexylmethylguanine derivatives: potent inhibitors of cyclin-dependent kinases 1 and 2. J Med Chem 47:3710–3722. https://doi.org/10.1021/jm0311442
Gutiérrez-de-Terán H, Sallander J, Sotelo E (2017) Structure-based rational design of adenosine receptor ligands. Curr Top Med Chem 17:40–58. https://doi.org/10.2174/1568026616666160719164207
Bjelic S, Brandsdal BO, Åqvist J (2008) Cold adaptation of enzyme reaction rates. Biochemistry 47:10049–10057. https://doi.org/10.1021/bi801177k
Azuaje J, Jespers W, Yaziji V et al (2017) Effect of nitrogen atom substitution in A3 adenosine receptor binding: N-(4,6-diarylpyridin-2-yl)acetamides as potent and selective antagonists. J Med Chem 60:7502–7511. https://doi.org/10.1021/acs.jmedchem.7b00860
Liu W, Chun E, Thompson AA et al (2012) Structural basis for allosteric regulation of GPCRs by sodium ions. Science 337:232–236. https://doi.org/10.1126/science.1219218
Jespers W, Schiedel AC, Heitman LH et al (2018) Structural mapping of adenosine receptor mutations: ligand binding and signaling mechanisms. Trends Pharmacol Sci 39:75–89
Böhm HJ, Flohr A, Stahl M (2004) Scaffold hopping. Drug Discov Today Technol 1:217–224. https://doi.org/10.1016/j.ddtec.2004.10.009
Wang L, Deng Y, Wu Y et al (2017) Accurate modeling of scaffold hopping transformations in drug discovery. J Chem Theory Comput 13:42–54. https://doi.org/10.1021/acs.jctc.6b00991
Authors’ contributions
WJ and HGT designed the workflow and the calculations; WJ and ME programmed the workflow; WJ performed the calculations; WJ, JÅ and HGT analysed the results; WJ, JÅ and HGT wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Christoffer Lind for helpful discussions.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
QligFEP is freely available at: https://github.com/qusers/qligfep.
Funding
This study was funded by the Swedish Research Council (VR). Additional support from the Swedish strategic research programme eSSENCE is acknowledged. The computations were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jespers, W., Esguerra, M., Åqvist, J. et al. QligFEP: an automated workflow for small molecule free energy calculations in Q. J Cheminform 11, 26 (2019). https://doi.org/10.1186/s13321-019-0348-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13321-019-0348-5